Species library
Scope
The user-defined species library is a core component to photoQuad's functionality, allowing the association of various object properties to particular species. The library can be an MS Excel (*.xls) or a typical comma-separated ASCII file (*.csv), and it can be updated with new species entries in real-time, during data analysis and exploration.
Highlights
Choice between two different file formats (MS Excel or plain ASCII)
Multiple species libraries supported per user
The library can be located at any particular directory
Filter and sorting tools available
Dedicated tools to add/remove entries during analysis
Association of multiple preview images to particular library entries
photoQuad ships with an embedded demo library to get you started
Read more: Library format | The species library GUI | Add/remove entries | Preview images | FAQs
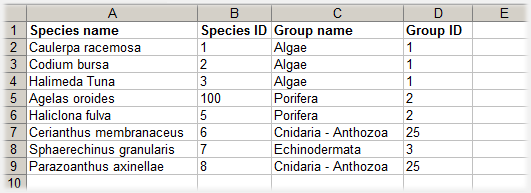
The species library is a 4-column file with the following entries, in the order specified:
| Column header | Format | Example entry |
| Species name | text string | Axinella cannabina |
| Species ID | integer >0, unique per Species (regardless Group) | 101 |
| Group name | text string | Porifera |
| Group ID | integer >0, unique per Group | 2 |
Each species entry is represented by a single row, as shown in the example below:
You can use any Species name, Species ID, Group name or Group ID, under the following rules:
Each Species (1st column) must have a unique numeric Species ID (2nd column).
A Species ID is globally unique, i.e. it belongs to one species only and cannot reappear anywhere again in the 2nd column.
Species are lumped together under a Group (3rd column). Again, each Group has a unique Group ID (4th column).
Species and Group IDs do not have to be consecutive; they just need to be positive integers and unique within the Species and Group columns.
There is no conflict if a Species has the same numeric ID with a Group.
There is no limit in the number of library entries (rows).
Blank rows, missing data (e.g. no Group name), and non-English characters are not allowed.
photoQuad can use a species library saved either as an MS Excel file (*.xls) or as a simple comma-separated ASCII file (*.csv). In the second case, the same file shown in the above worksheet would look like this in a text editor:

WARNING:
An ill-formated species library will cause unpredictable photoQuad behavior and erroneous results. The most common mistake is the use of duplicate Species IDs in the 2nd column.
NOTE:
You must have MS Excel installed (or LibreOffice/OpenOffice) in order to use a species library in Excel format (*.xls). photoQuad will still be able to read/write *.xls files, but you won't be able to view or edit them.
If you don't have MS Excel installed (or an equivalent software that can edit *.xls files), you should only use the plain ASCII text (*.csv) option, which is better anyway...
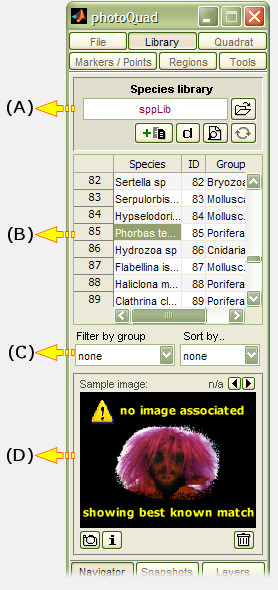
The species library has its own dedicated tab panel in photoQuad's interface.
(A) File panel
The current library filename is displayed here, alongside with buttons for: File browsing, Refresh library, Show library expected format, Load embedded demo library, and add/remove species entry.
(B) Library display
This is a simple Table where library contents are displayed. The Table fields are non-editable but interactive: clicking on a species entry displays the associated images in field (D), if any.
(C) Sorting tools
The species library can become quite lengthy, and these filter and sorting tools facilitate its management. Entries can be filtered by Group, or sorted by A..Z - Species, A..Z - Group, Z..A - Species, Z..A Group, or by Species ID.
(D) Preview image association panel
This tool allows a custom image region, or multiple regions from different images, to be associated to a particular library entry. The next time you click on a library entry, the associated preview images are displayed, including the source image metadata. Read more about this feature here.
Being a simple spreadsheet or comma-separated ASCII file, the species library can be easily viewed and modified manually, without using photoQuad. Edit the *.xls file in a spreadsheet editor (or the *.csv file in a text editor), save it, and press the "Refresh library" button.
As the file grows bigger however, its manual management gets harder, and chances are that spelling mistakes will find their way into the library, or even worse, duplicate Species and Group IDs may be entered. To circumvent these issues, photoQuad is equipped with a dedicated interface for adding species library entries during runtime, with automatic entry validation checks.
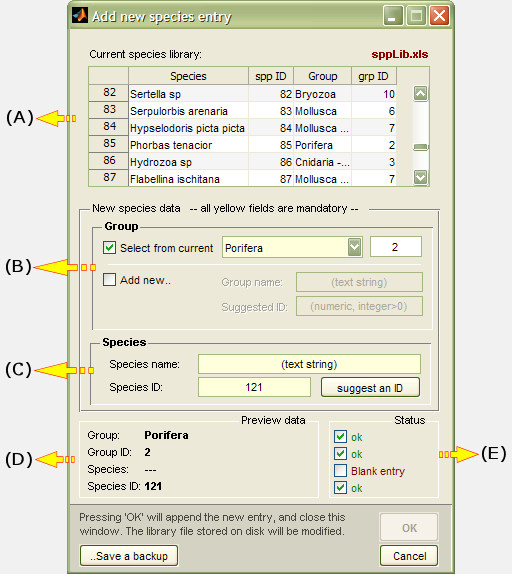
(A) Library display
A non-editable field where the current library contents are displayed.
(B) Add new entry: Group
Each Species entry must be associated with a more general category, i.e. a "Group", which is used to cluster similar species together and facilitate their quick search and allocation to analysis objects. Each Group must have a unique identification number (the Group ID), and photoQuad offers two entry options.
Select from current: An entry is selected out of a list of pre-existing Groups. Both Group name and Group ID are thereby automatically assigned.
Add new: A new Group name is manually entered by the user, and photoQuad automatically suggests a valid and unique new Group ID, having scanned the existing library. The suggested Group ID can be replaced by a custom one, but photoQuad will have the final word on this: if the custom ID is already assigned to another Group, an auto-correction is applied.
(C) Add new entry: Species
A new Species name is manually entered by the user, and photoQuad automatically suggests a valid and unique new Species ID, having scanned the existing library for conflicts. Auto-correction checks apply exactly as described in (B).
(D) New entry preview pane
Displays a preview of summary of the new library entry. Non-editable.
(E) New entry validation report
During the whole process, photoQuad checks the input data for potential conflicts (duplicate IDs, case-sensitive differences, etc) and updates the validation report in real time. A new entry cannot be submitted unless all checkboxes are green (OK).
Species preview images can help as a visual reference during analysis, and can be saved the moment new species are identified. photoQuad allows the selection of a custom image region, or regions from different images, to be associated to a particular library entry during analysis. It only takes a few clicks, and the illustrations below show how it's done:
1) Select a library entry from photoQuad's main GUI.
2) Click the button with the camera icon, and a draggable/adjustable rectangle appears on the image.
3) Select the desired image region and press "Return". Done.
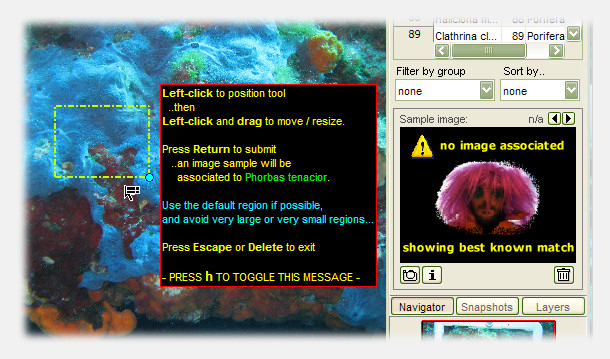
Multiple preview images can be associated to single library entry, and source image metadata are also stored, so that preview images can be traced back to their origin.
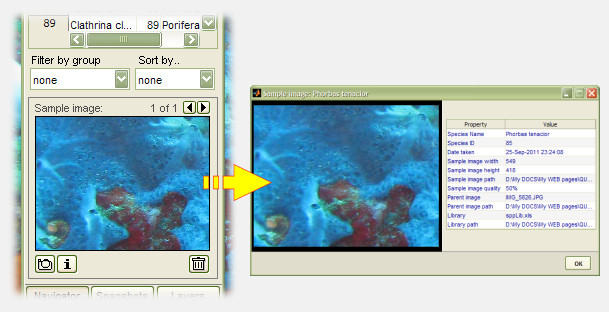
NOTE:
The preview images are actually associated with the corresponding Species ID; therefore, they are currently only supported for one species library. This means that you can use multiple species libraries for your various analyses, but can only associate preview images to one of them. We will fix this in an upcoming photoQuad version.
Create a backup of your preview images
This is useful when you want to import your preview images after a new photoQuad installation or update, or you just want to make a safe copy of them.
Go to the "Library" menu, and select "Create backup of species preview images". Store them somewhere safe, and you can browse them there. Do not rename these files if you intend to re-import them to photoQuad.
Use the respective "Import backup" on the "Library" menu to re-import them to photoQuad. Note that the imported backup will replace the currently used preview images for that particular photoQuad installation.
Is there a maximum number of library entries?
No, but note that if you use an MS Excel file, you are limited by the number of lines the Excel software allows.
Why do you support two different file formats?
So that MS Office is not needed in order to use photoQuad. Next to that, comma-separated ASCII files are cooler and much more manageable.
What happens if I have already defined some analysis objects (e.g. regions, random points, etc) but forgot to import a species library?
No worries. photoQuad is designed to make these things easy to forget. Try to associate an object to a species: if a library is there, things will work as normal; if no library has yet been imported, photoQuad will prompt for one or suggest to use the embedded library. The work done on analysis objects is not affected.
What happens if I enter duplicate Species or Group IDs?
You severely compromise your results, and you are up for unpredictable photoQuad behaviour. Please note that Species names and Group names are only there so that we, earthlings, can understand what's going on. photoQuad only cares for those numeric IDs, and it expects them to be unique per Species and Group category. Therefore, it is always safer to use photoQuad's interface to add/remove library entries.
Do Species or Group IDs need to be consecutive?
No, they just need to be positive integers and unique per Species and Group category. If you had a library with 4 species, their IDs could be "1, 2, 100, 3" or "1, 2, 3, 4", but not "-1, 2, -100, 3" or "1, 1, 2, 1". It's that simple.
How do I start a new species library?
You can do this in multiple ways:
Method 1: download this file (*.xls) or this one (*.csv), and use it as a template. Leave the first line untouched (Headers), delete all entries from line 2 and downwards, read the simple specifications described on this page, and start using your custom library.
Method 2: Launch photoQuad and import the embedded demo library. To do this, either press the "Load demo species library" button found on the "Library" tab, or select "Import demo library" from the image's menu. Now press the "Add/remove entries" button found on the "Library" tab. On the new window, you can find a "..Save a backup" button at the bottom-left corner. Use it to make a copy of the embedded library, and start modifying as described on Method 1 above.
Can I add more columns to my species library file (for notes, temporary entries etc)?
Yes. photoQuad only reads the first 4 columns and ignores the rest.
Are blank rows allowed in the species library file?
No. This will result to a "blank" Species name, but there will be no numeric ID for it, so photoQuad will be confused.
Can I remove the Headers from my species library file?
No.
Do I have to spell the Headers exactly as they are in the demo libraries?
No, but why would you want to change this? Please treat the species library as a sensitive system file. The whole photoquadrat analysis process can be rendered useless if the species library is messed up, so best practice is to start building your library using the demo files provided. Please avoid improvisations here.
How can I access my species preview images?
Go to the "Library" menu, and select "Create backup of species preview images". Store them somewhere safe, and you can browse them there. Do not rename these files if you intend to re-import them to photoQuad, e.g. after a new photoQuad update installation.